Major Projects
1) Bacterial polysaccharide biosynthesis systems are a key interest in our group with an emphasis on developing in vitro systems for the enzymatic synthesis of these complex polymers. The goal of this series of projects is to develop libraries of well-characterized enzymes for the assembly of these materials. The eventual application of this work will be to have methods to produce any bacterial polysaccharide for biomedical applications such as therapeutics based on symbiotic bacterial capsules or vaccines against pathogenic bacteria. In addition, this work will eventually focus on the biophysical properties of these complex polysaccharide materials to understand their diverse ranges of bioactivity. Work in this area typically involves techniques in molecular biology, protein expression, isolation and purification, and the use of bioanalytical techniques such as Capillary Electrophoresis and High Performance Liquid Chromatography. Typical organic characterization techniques are also utilized including Mass Spectral identification of products and multidimensional NMR techniques.
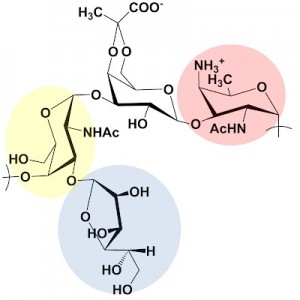
The above polysaccharide is the repeating unit of the capsule structure from the organism Bacteroides fragilis. This sugar has been shown to have important effects on normal development of the mammalian immune system and is one of several targets for the Troutman group.
2) Bacterial polysaccharide biosynthesis probes will be and have been developed in our group for characterizing the assembly of these molecules. The goal of projects in this area is to more easily track the biosynthesis of complex polysaccharide polymers on the isoprenoid scaffold bactoprenyl phosphate. Our interest here is in developing both in vitro and in cellulo probes for these assembly pathways to identify enzymes responsible for their biosynthesis. To do this we use tools in organic chemistry, chemical biology and enzymology. The ultimate goal of this area of research is to design novel probes for characterizing these materials to identify potential targets for anti-bacterial agents.
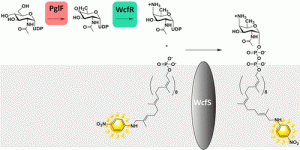
The above image is the highlight figure from our recent article in the journal Biochemistry, where we utilized a chromophore containing bactoprenyl phosphate to characterize the first step in the assembly of the polysaccharide capsule of Bacteroides fragilis Capsular Polysaccharide A.
3) Undecaprenyl Pyrophosphate Synthase (UPPS) is a critical enzyme for bacterial survival. Our goal in this series of projects is to understand how UPPS from different species recognize different substrate structural motifs. With knowledge of these differences we will begin the development of new species selective inhibitors for UPPS. The work in these projects will utilize techniques in organic chemistry, molecular biology, site-directed mutagenesis, protein expression, isolation and purification, as well as method development in rapid assay design and HPLC methodology. We have also established a strong collaboration with Dr. Donald Jacobs of the UNC-Charlotte Physics Department for the development of computational tools to study this protein.
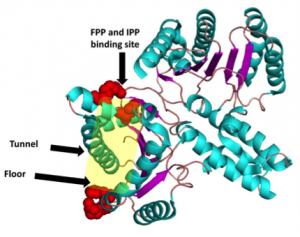
The above image is a representative structure of a UPPS protein highlighting the key areas for substrate interactions and elongation of bactoprenyl diphosphate.
4) Developing protein-lipid hybrid building blocks for nanoscale science applications. A major focus of the UNC-Charlotte Chemistry department is in Nanoscale Science. Our work in polysaccharide biosynthesis will eventually move in that direction, but we have also recently begun developing a research program that focuses on the development of self assembling building blocks with proteins embedded in lipid-protein nanoparticles called nanodiscs (nanodiscs were first developed by the Sligar group at the University of Illinois Urbana-Champagne). The goal of projects in this area is to assemble protein specific biomachines taking advantage of protein-protein interactions well conserved in nature. The eventual applications of these materials could be in the development of nanofiltration systems, reaction centers, and metamaterials.
Joining the Laboratory:
If you are interested in working with me contact me at Jerry.Troutman@uncc.edu. I take care to fill my lab with only people that are truly interested in science and research. Research is an immensely rewarding experience, but only with plenty of sweat. Being smart is not enough to be successful. You should be ready to be challenged at all times, and you should thrive under pressure. Medicine exists because of what researchers do every day. Technology exists because of what researchers do everyday. If you want to be a part of that, then I can’t wait to meet you.
Undergraduate Students:
If you are an undergraduate interested in my laboratory I expect dedication and commitment to research. You should be ready to work hard and learn a ton. If you are signing up for credit I only accept three and four credit hour commitments. I expect you to desire to be independent and be capable of becoming so rapidly. I expect you to perform original research, read the literature, attend group meetings, and contribute intellectually to our work.
Graduate Students:
Graduate school is challenging, requiring you to manage time like you have never had to do before. You must be able to juggle many things at once and the fact of the matter is: that is part of your training. I expect my graduate students to focus very hard on publications, and nearly all work should have publication in mind at all times. I expect you to be a model for the undergraduates in the laboratory, and help in the training of these undergraduates, as this is also part of your training. If you are interested please take a look at both the Chemistry Masters and Nanoscale PhD program. In addition, there is an interdisciplinary PhD program in Biology. I am always happy to work with students interested in biomedical and biochemical research, and am committed to making you the best researcher that you can be. Our lab is also affiliated with The Center for Biomedical Engineering Systems, which you should also take a look at.