Study Sites
I. Transmission dynamics of malaria parasites
+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Human movements and dispersal of vector mosquitoes allow spread of malaria and impact genetic structure of the parasite populations. Understanding the relatedness among Plasmodium populations and how such influenced by geographical and environmental features is vital. The combined approach of population genomics and landscape genetics allows us to examine the role of spatial variation and local adaptation in disease transmission. Knowledge of the source/sink of infections and the magnitude of disease spread are keys to make decision and implement control measures effectively in the most malarious areas.
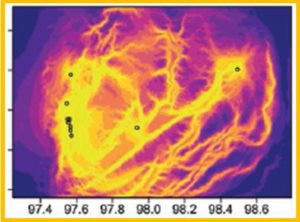
Landscape genetic plot showing the level of gene flow among P. vivax populations in the Myanmar-China border area.
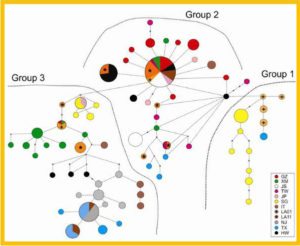
Bayesian inferences of genetic clusters by STRUCTURE among P. vivax samples in Myanmar.
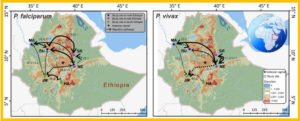
Migratory pathways and rates of Plasmodium falciparum and P. vivax in Ethiopia based on Migrate-N and BayesAss.
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
II. Molecular epidemiology of malaria
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Emergence of antimalarial drug resistance and persistence of submicroscopic parasite reservoirs are apparent hurdles to malaria reduction and elimination in many African countries. These issues require close monitoring and effective resolution. One aspect of my research is to examine demographic factors of malaria prevalence, transmission intensity, disease severity, as well as bio-markers related to antimalarial drug resistance. This information will contribute to assessing and monitoring the disease burden in places where people are still battling or at high risk of malaria.
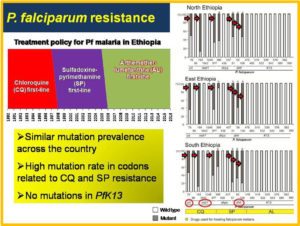
Antimalarial treatment policy in Ethiopia and mutation prevalence of antimalarial resistance genes in Plasmodium falciparum.
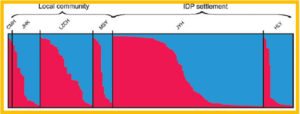
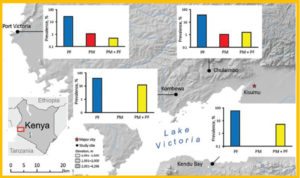
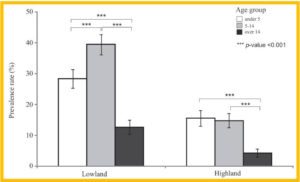
Histogram showing the mean malaria prevalence rate of the three age groups (under 5, aged 5–14, and over 14) in the lowlands and highlands of Western Kenya.
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
III. Evolution of parasite-host interactions
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Malaria is caused by the invasion of Plasmodium into human red blood cell and subsequently disrupts its normal function. RBC invasion involves multiple interactions between parasite ligand and host receptor proteins. Although a number of invasion ligands have been recently documented, in very few cases have their functional significance and red blood cell receptors been identified, especially in P. vivax. It is always possible that parasites evolve novel RBC invasion pathways. This could influence the efficacy of existing preventive vaccines and elevate malaria burden at a global level. My research explores molecular mechanisms of parasite invasion with the goal to identify key RBC binding proteins.
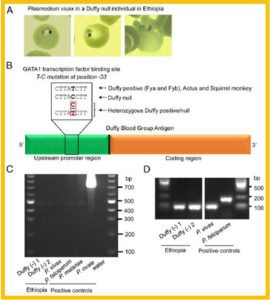 Detection of P. vivax infection in Duffy-negative Ethiopians by microscopy and PCR method.
Detection of P. vivax infection in Duffy-negative Ethiopians by microscopy and PCR method.
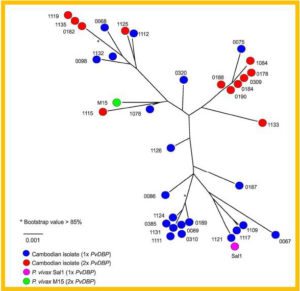
High copy number of PvDBP observed in Plasmodium vivax from Duffy negative patients in Ethiopia based on quantitative real-time PCR.
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
IV. Phylogeny of malaria parasites and vectors
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Genetic relationships among biological organisms take into account the mutational changes occurred over time as well as migration rates. My research examines mutational and genotypic variation within parasite species and explores genetic relatedness at the spatial and temporal scales to elucidate ancestral origins and character changes. This involves the use of geo-reference tools to model geological changes with respect to species movement and distribution over time. This information sheds light on the role and impact of long-distance migration and adaptive radiation on species diversity and population structure.
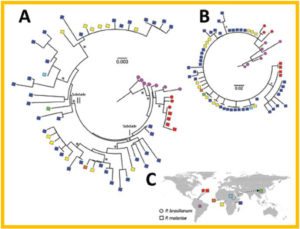 Maximum-likelihood analyses of circumsporozoite protein gene (csp) sequences of Plasmodium malariae and global distribution of the samples.
Maximum-likelihood analyses of circumsporozoite protein gene (csp) sequences of Plasmodium malariae and global distribution of the samples.
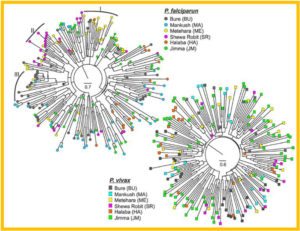
Neighbor-joining trees showing the genetic relatedness among Plasmodium falciparum and P. vivax samples in Ethiopia.