Shan Yan, PhD, Professor
Research Interests: Molecular Mechanisms of Genome Integrity and Cancer Etiology
Research Areas: Cancer Biology, Cell Biology, Molecular Biology, Developmental Biology, Environmental Health, Xenopus laevis
Lab website:
Dr. Yan:
Dr. Shan Yan is Professor in the Department of Biological Sciences at UNC Charlotte. After postdoctoral/research training in the Department of Molecular and Cellular Biology at Harvard University, he joined the faculty of UNC Charlotte as tenure-track Assistant Professor in 2010 and was promoted to Associate Professor (with tenure) in 2016 and Full Professor in 2019. Since 2017, Dr. Yan has initiated and directed the Charlotte Biology and Biotechnology (CBB) Exchange Group co-sponsored by North Carolina Biotechnology Center (NCBC) and UNC Charlotte. He is also a Program Leader of the Genome Integrity and Cancer Initiative (GICI) at UNC Charlotte.
Dr. Yan has established a productive research program working on genome integrity and cancer etiology, which has been supported by funds from UNC Charlotte, and grants from external funding agencies including NCBC and the NIH (NCI, NIEHS, and NIGMS) through R01, R21, R03, and R15 mechanisms. His dedicated quality teaching and mentoring is evidenced by effective classroom teaching of graduate and undergraduate courses and an extensive mentoring experience for 2 junior tenure-track faculty members, 3 Research faculty/staff, 4 postdoctoral fellows, 13 graduate students, and 26 undergraduate students including several women students and those from underrepresented minority groups.
Dr. Yan’s achievements in research and teaching have been recognized by several honors and awards. He is the recipient of the 2023 EMGS Education Award from the Environmental Mutagenesis & Genomics Society (EMGS). Dr. Yan is granted the 2022 Outstanding Data Science Faculty Research Award from School of Data Science at UNC Charlotte. Dr. Yan also served on several leadership roles such as Director on the Board of Directors of the Federation of American Society For Experimental Biology (FASEB), Chair of EMGS Awards & Honors Committee, Chair of EMGS Publication Policy Committee, and Chair of the Competitive Grant Committee in the University Faculty Council at UNC Charlotte.
Dr. Yan has served as grant reviewer for several NIH Study Sections and NSF Review Panels. He has also been Associate Editor, Managing Editor or Editorial Board Member for several peer-reviewed journals including Journal of Biological Chemistry, Environmental and Molecular Mutagenesis, Computational and Structural Biotechnology Journal, Plos ONE, and Frontiers in Cell and Developmental Biology. Dr. Yan has been manuscript reviewer for >50 leading scientific journals such as Science, Trends in Cell Biology, Nature Communications, Nucleic Acids Research, PNAS, Cancer Research, eLife, Cell Reports, Molecular Biology of the Cell, and Journal of Biological Chemistry.
Yan Lab Research:
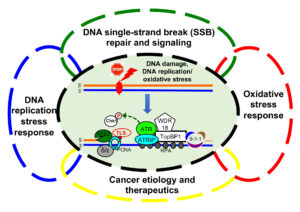
The Yan lab focuses on the molecular mechanisms of genome integrity and cancer etiology. Genome of eukaryotes are constantly exposed to threats from endogenous sources such as oxidative stress and exogenous sources such as environmental agents. In response to DNA damage or stressful conditions, cells have evolved a variety of DNA repair and DNA damage response (DDR) pathways to maintain genome stability. Defective DNA repair and DDR pathways have been found in associated with human diseases, such as cancer, sepsis, aging, and neurodegenerative disorders. Using bioinformatics, biochemical, biophysical, molecular and cell biology approaches, the Yan lab is interested in various significant aspects of genome integrity maintenance using Xenopus egg extracts and mammalian cell lines as model systems. Anticipated findings from various research programs/projects will help to better understand how cells maintain genome stability and to provide novel avenues for cancer diagnostics and therapeutics.
The Yan laboratory has made several contributions to science:
(1) Dissecting DNA single-strand break repair and ATR-/ATM-mediated checkpoint signaling pathways (Lin et al., Nucleic Acids Res, 2018; Yan et al., Nat Struct Mol Biol, 2019; Lin et al., Nucleic Acids Res, 2020; ; Zhao et al., Nature Communications, 2024);
(2) Elucidating molecular mechanisms of DNA damage response pathway in oxidative stress (Willis et al., PNAS, 2013; Yan et al., Cell Mol Life Sci, 2014; Wallace et al., PNAS, 2017; Cupello et al., Biochem J, 2019);
(3) Revealing the molecular mechanism of maintaining ssDNA stability (Bai et al., Cell Signal, 2014; Ha et al., J Biol Chem, 2020; Lin et al., eLife, 2023);
(4) Demonstrating novel regulatory mechanisms of nucleolar DNA damage response by liquid-liquid phase separation and biomolecular condensates (Li et al., Nucleic Acids Res, 2022; Li and Yan, Trends Cell Biol, 2023);
(5) Characterizing novel basic mechanisms of cancer etiology and therapeutics (Li et al., Cancer Res, 2017; Jensen et al., Sci Rep, 2020; Hu et al., Cancer Res, 2021; Hossain et al., Front Cell Dev Biol, 2021; McMahon et al., NAR Cancer, 2023).
Research Directions/Projects:
(1) To decipher the regulation and crosstalk of SSB/DSB repair and signaling pathways
(2) To elucidate the molecular mechanisms of nucleolar and mitochondrial DNA/RNA stability
(3) To dissect the molecular details of biomolecular condensates (liquid-liquid phase separation) in genome integrity
(4) To modulate the mechanisms of APE1, and APE2, and PARP1 in genome integrity for the treatment of BRCA1-/BRCA2-related cancers
Representative Publications (*corresponding author):
(1) Willis J, Patel Y, Lentz B, Yan S*. 2013. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. PNAS. 110 (26): 10592-10597.
(2) Yan S*, Sorrell M, Berman Z. 2014. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cellular and Molecular Life Sciences. 71 (20): 3951-3967.
(3) Wallace BD, Berman Z,, Mueller GA, Lin Y, Chang T, Andres SN, Wojtaszek JL, DeRose EF, Appel CD, London RE, Yan S*, Williams RS*. 2017. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. PNAS. 114 (2):304-309.
(4) Lin Y, Bai L, Cupello S, Hossain MA, Deem B, McLeod M, Raj J, Yan S*. 2018. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Research. 46 (5): 2479-2494.
(5) Yan S*. 2019. Resolution of a complex crisis at DNA 3′ termini. Nature Structural & Molecular Biology. 26 (5): 335-336.
(6) Ha A, Lin Y, and Yan S*. 2020. A non-canonical role for the DNA glycosylase NEIL3 in suppressing APE1 endonuclease–mediated ssDNA damage.Journal of Biological Chemistry. 295 (41): 14222-14235.
(7) Lin Y, Raj J, Li J, Ha A, Hossain MA, Richardson C, Mukherjee P, Yan S*. 2020. APE1 senses DNA single-strand breaks for repair and signaling. Nucleic Acids Research. 48(4):1925-1940.
(8) Li J, Zhao H, McMahon A, . 2022. APE1 assembles biomolecular condensates to promote the ATR-Chk1 DNA damage response in nucleolus. Nucleic Acids Research. 50(18):10503-10525.
(9 Li J, Yan S. 2023. Molecular mechanisms of nucleolar DNA damage checkpoint response. Trends in Cell Biology. 33 (5): 361-364.
(10) Lin Y, Li J, Zhao H, McMahon A, McGhee K, . 2023. APE1 recruits ATRIP to ssDNA in an RPA-dependent and -independent manner to promote the ATR DNA damage response. eLife. 12: e82324.
(11) McMahon A, Zhao J, Yan S*. 2024. Ubiquitin-mediated regulation of APE2 protein abundance. Journal of Biological Chemistry. 300 (6): 107337.
(12) Zhao H, Li J, You Z, Lindsay HD, Yan S*. 2024. Distinct regulation of ATM signaling by DNA single-strand breaks and APE1. Nature Communications. 15 (1): 6517.
Click the link for the video (~10 minutes):
JoVE-Video
Willis J, DeStephanis D, Patel Y, Gowda V, and Yan S*. 2012. Study of the DNA damage checkpoint using Xenopus egg extracts. Journal of Visualized Experiments. (69): e4449 10.3791/4449. DOI: http://dx.doi.org/10.3791/4449
Click the link for the webinar by Dr. Yan to the NIH DNA Repair Interest Group (~65minutes):
Videocast at NIH.
Department of Biological Sciences website:https://biology.charlotte.edu/
College of Science website: https://science.charlotte.edu/
ORCID: https://orcid.org/0000-0001-8106-6295
QR code for the Yan lab website