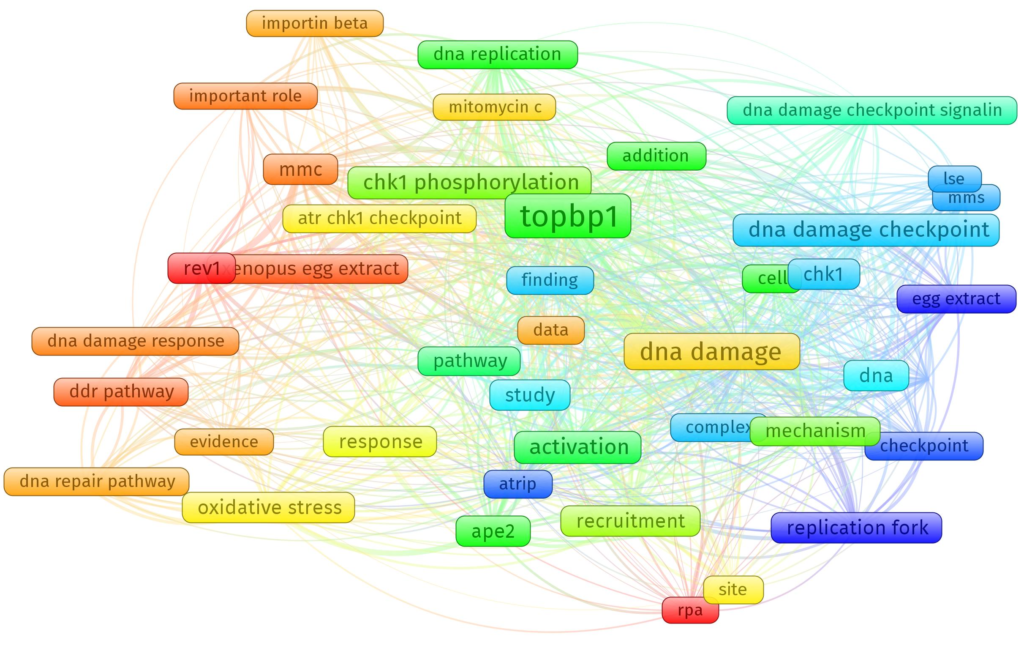
Notes:
§: equal contribution
‡: undergraduate student
¶: graduate student
#: postdoctoral fellow/research associate/research faculty
*: corresponding author
>>>: preprint
Publications after joining UNC Charlotte (total=n)
2025
>>>55. McMahon AM¶, Zhao H#,Li J, Driscoll G¶, Matos J¶, McGhee K¶, Lyttle J‡, Yan S*. 2025. PARP1 promotes replication-independent DNA double-strand break formation after acute DNA-methylation damage. bioRxiv [Preprint]. 2025 Jul 12: 2025.07.10.663928. (PMCID: PMC12265532; PMID: 40672314) DOI: https://doi.org/10.1101/2025.07.10.663928.
2024
54. Zhao H#, Richardson C, Marriott I, Yang IH, Yan S*. 2024. APE1 is a master regulator of the ATR-/ATM-mediated DNA damage response. DNA Repair. 144: 103776. (Published online October 19, 2024) (PMCID: PMC11611674; PMID: 39461278) DOI: https://doi.org/10.1016/j.dnarep.2024.103776
53. Zhao H#, Li J#, You Z, Lindsay HD, Yan S*. 2024. Distinct regulation of ATM signaling by DNA single-strand breaks and APE1. Nature Communications. 15 (1): 6517. (Published online August 7, 2024) (PMCID: PMC11306256; PMID: 39112456) DOI: https://doi.org/10.1038/s41467-024-50836-6
♪ Public news release 08/07/2024. “Demystifying APE1: New findings on direct activation of ATM signaling by DNA single-strand breaks” (Please see the link @ Eurekalert)
52. McMahon A¶, Zhao J, Yan S*. 2024. Ubiquitin-mediated regulation of APE2 protein abundance. Journal of Biological Chemistry. 300 (6): 107337. (PMCID: PMC11157268; PMID: 38705397) DOI: https://doi.org/10.1016/j.jbc.2024.107337
51. Yan S*, 2024. Inspiring basic and applied research in genome integrity mechanisms: Dedication to Samuel H. Wilson. Environmental and Molecular Mutagenesis. 65 (S1): 4-8. (PMCID: PMC11110888; PMID: 38619433) DOI: https://doi.org/10.1002/em.22595
50. Suptela AJ, Radwan Y, Richardson C, Yan S, Afonin KA, Marriott I*. 2024. cGAS mediates the inflammatory responses of human microglial cells to genotoxic DNA damage. Inflammation. 47 (2): 822-836. (PMCID: PMC11073916; PMID: 38148453) DOI: https://doi.org/10.1007/s10753-023-01946-8
2023
49. Driscoll G¶, Yan S*. 2023. Maintaining chromosome and genome stability by the RPA-RNF20-SNF2H cascade. Proceedings of the National Academy of Sciences of the United States of America. 120 (24): e2306455120. (PMCID: PMC10268305; PMID: 37276422). DOI: https://doi.org/10.1073/pnas.2306455120
48. Lin Y#, Li J#, Zhao H#, McMahon A¶, McGhee K¶, . 2023. APE1 recruits ATRIP to ssDNA in an RPA-dependent and -independent manner to promote the ATR DNA damage response. eLife. 12: e82324. (PMCID: PMC10202453; PMID: 37216274) DOI: https://doi.org/10.7554/eLife.82324
47. Li J#, Yan S. 2023. Molecular mechanisms of nucleolar DNA damage checkpoint response. Trends in Cell Biology. 33 (5): 361-364. (Online published on March 16, 2023) (PMCID: PMC10215988; PMID: 36933998) DOI: https://doi.org/10.1016/j.tcb.2023.02.003
♪ Public release 03/16/2023. “Researchers highlight nucleolar DNA damage response in fight against cancer” (Please see the link @ Eurekalert or @ UNC Charlotte )
46. McMahon A¶, Zhao J, . 2023. APE2 catalytic function and synthetic lethality draw attention as a cancer therapy target. NAR Cancer. 5 (1):zcad006. (PMCID: PMC9900424 ; PMID: 36755963) DOI: https://doi.org/10.1093/narcan/zcad006
2022
45. Li J#, Zhao H, McMahon A¶, . 2022. APE1 assembles biomolecular condensates to promote the ATR-Chk1 DNA damage response in nucleolus. Nucleic Acids Research. 50(18):10503-10525. (PMCID: PMC9561277; PMID: 36200829) (online published October 6, 2022). DOI: https://doi.org/10.1093/nar/gkac853
♪ Public release 03/16/2023. “Researchers highlight nucleolar DNA damage response in fight against cancer” (Please see the link @ Eurekalert or @ UNC Charlotte )
44. , Zhao J*, Kemp M*, Sobol RW*. 2022. Editorial: mechanistic studies of genome integrity, environmental health, and cancer etiology. Frontiers in Cell and Developmental Biology. 10: 1026326. (PMCID: PMC9554606; PMID: 36247007) DOI: https://doi.org/10.3389/fcell.2022.1026326
>>>43. Li J#, Zhao H#, McMahon A¶, . 2022. APE1 assembles biomolecular condensates to promote the ATR-Chk1 DNA damage response in nucleolus. bioRxiv. (preprint online August 22, 2022) DOI:
>>>42. Lin Y, Li J#, Zhao H#, McMahon A¶, . 2022. APE1 recruits ATRIP to ssDNA in an RPA-independent manner to promote the ATR DNA damage response. bioRxiv. (preprint online August 12, 2022) DOI: https://doi.org/10.1101/2022.08.12.503732
41. Sadaf H, Hong H, Maqbool M, Emhoff K, Lin J, Anwer F, and Zhao J. 2022. Multiple myeloma etiology and treatment. Journal of Translational Genetics and Genomics. 6:63-83. (online ahead of print January 20, 2022) DOI: https://doi.org/10.20517/jtgg.2021.36
40. Tarannum M, Hossain MA, Holmes B, Mukherjee P, Vivero-Escoto JL. 2022. Advanced nanoengineering approach for target-specific, spatiotemporal, and ratiometric delivery of gemcitabine-cisplatin combination for improved therapeutic outcome in pancreatic cancer. Small. 18(2):e2104449. (online ahead of print November 10, 2021). (PMCID: PMC8758547; PMID: 34758094) DOI: https://doi.org/10.1002/smll.202104449
2021
39. Hossain MA¶, Lin Y#, Driscoll G¶, Li J#, McMahon A¶, Matos J¶, Zhao H#, Tsuchimoto D, Nakabeppu Y, Zhao J, Yan S*. 2021. APE2 is a general regulator of the ATR-Chk1 DNA damage response pathway to maintain genome integrity in pancreatic cancer cells. Frontiers in Cell and Developmental Biology. 9:738502. (PMCID: PMC8593216; PMID: 34796173) DOI: https://doi.org/10.3389/fcell.2021.738502
38. Huff LA, , and Clemens MG. 2021. Mechanisms of Ataxia Telangiectasia Mutated (ATM) control in the DNA damage response to oxidative stress, epigenetic regulation, and persistent innate immune suppression following sepsis. Antioxidants. 10 (7): 1146. (PMCID: PMC8301080; PMID: 34356379) DOI: https://doi.org/10.3390/antiox10071146
37. . 2021. Cisplatin-mediated upregulation of APE2 binding to MYH9 provokes mitochondrial fragmentation and acute kidney injury. Cancer Research (online ahead of print December 7, 2020). 81 (3): 713-723. (PMCID: PMC7869671; PMID: 33288657) DOI: https://doi.org/10.1158/0008-5472.CAN-20-1010
36. Lin Y#, McMahon A¶, Driscoll G¶, Bullock S, Zhao J, and Yan S*. 2021. Function and molecular mechanisms of APE2 in genome and epigenome integrity. Mutation Research-Reviews in Mutation Research (online ahead of print November 16, 2020). 787 (January–June): 108347. (PMCID: PMC8287789; PMID: 34083046) DOI: https://doi.org/10.1016/j.mrrev.2020.108347
2020
35. Ha A¶, Lin Y#, and Yan S*. 2020. A non-canonical role for the DNA glycosylase NEIL3 in suppressing APE1 endonuclease-mediated ssDNA damage. Journal of Biological Chemistry (online ahead of print August 14, 2020). 295 (41): 14222-14235. (PMCID: PMC7549043; PMID: 32817342) DOI: https://doi.org/10.1074/jbc.RA120.014228
♪Prakash A and Sharma N: Faculty Opinions Recommendation of [Ha A et al., J Biol Chem 2020 295(41):14222-14235]. In Faculty Opinions, 16 Dec 2020; DOI: http://doi.org/10.3410/f.738528228.793580924
34. Yan S* and Vaziri C*. 2020. An introduction for the special issue on environmental health and genome integrity. Environmental and Molecular Mutagenesis., 61 (7): 660-663. (PMCID: PMC7442621; PMID:32683747; online ahead of print July 19, 2020) DOI: https://doi.org/10.1002/em.22400
33. Jensen KA¶, Shi X, Yan S*. 2020. Genomic alterations and abnormal expression of APE2 in multiple cancers. Scientific Reports, 10 (1): 3758. (PMCID: PMC7048847; PMID:32111912) DOI: https://doi.org/10.1038/s41598-020-60656-5
>>>32. Jensen KA¶, Shi X, Yan S*. 2020. Genomic alterations and abnormal expression of APE2 in multiple cancers. bioRxiv (preprint online January 24, 2020) DOI: https://doi.org/10.1101/2020.01.17.910646
31. Yan S*., Williams C, and Huet Y. 2020. On the reproducibility of methods or findings. Lab Animal. 49 (29-31): 29. (PMCID: PMC7000164; PMID:31988408) DOI: https://doi.org/10.1038/s41684-019-0461-0
30. Lin Y§,#, Raj J§,‡, Li J#, Ha A¶, Hossain MA¶, Richardson C, Mukherjee P, Yan S*. 2020. APE1 senses DNA single-strand breaks for repair and signaling. Nucleic Acids Research. 48(4):1925-1940. (PMCID: PMC7038996; PMID: 31828326; Epub ahead of print) DOI: https://doi.org/10.1093/nar/gkz1175
♪Maiorano D: F1000Prime Recommendation of [Lin Y et al., Nucleic Acids Res 2019]. In F1000Prime, 03 Jan 2020; DOI: https://doi.org/10.3410/f.737076253.793568982
2019
29. Cupello S¶, Lin Y#, Yan S*. 2019. Distinct roles of XRCC1 in genome integrity in Xenopus egg extracts. Biochemical Journal. 476(24):3791-3804. (PMCID: PMC6959006; PMID: 31808793) DOI: https://doi.org/10.1042/BCJ20190798
28. Yan S*. 2019. Resolution of a complex crisis at DNA 3′ termini. Nature Structural & Molecular Biology. 26 (5): 335-336. (PMCID: PMC6549488; PMID: 30988507) DOI: https://doi.org/10.1038/s41594-019-0215-0
27. Lin Y#, Ha Anh¶‡, Yan S*. 2019. Methods for studying DNA single-strand break repair and signaling in Xenopus egg extracts. Methods in Molecular Biology: DNA Repair Methods and Protocols. 1999: 161-172. (PMCID: PMC6550457; PMID: 31127575) (Balakrishnan and Stewart eds). DOI: https://doi.org/10.1007/978-1-4939-9500-4_9
2016-2018
26. Hossain MA¶, Lin Y#, Yan S*. 2018. SSB end resection in genome stability: mechanism and regulation by APE2. International Journal of Molecular Sciences. 19 (8): 2389. (PMCID: PMC6122073; PMID:30110897) DOI: https://doi.org/10.3390/ijms19082389
25. Lin Y§,#, Bai L§,#, Cupello S¶, Hossain MA¶, Deem B¶, McLeod M¶, Raj J‡, Yan S*. 2018. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Research. 46 (5): 2479-2494. (PMCID: PMC5861430; PMID: 29361157) DOI: https://doi.org/10.1093/nar/gky020
24. Liu SY§, Li X§, Lin ZM, Su L, Yan S*, Zhao B*, Miao J*. 2018. SEC-induced activation of ANXA7 GTPase suppresses prostate cancer metastasis. Cancer Letters. 416: 11-23. (PMCID: PMC5777349; PMID: 29247827) DOI: 10.1016/j.canlet.2017.12.008
23. Li Z, Li J, Kong Y, Yan S, Ahmad N, Liu X*. 2017. Plk1 phosphorylation of Mre11 antagonizes DNA damage response. Cancer Research. 77(12): 3169-3180. (PMCID: PMC5504882; PMID: 28512243) DOI: 10.1158/0008-5472.CAN-16-2787
22. Wallace BD§, Berman Z§, ¶, Mueller GA, Lin Y#, Chang T, Andres SN, Wojtaszek JL, DeRose EF, Appel CD, London RE, Yan S*, Williams RS*. 2017. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 114 (2): 304-309. (PMCID: PMC5240719; PMID: 28028224) DOI: 10.1073/pnas.1610011114
♪ Public release by Hathaway, J. 01/06/2017. Study characterizes key molecular tool in DNA repair enzymes. (Please see the link @ Eurekalert or @ UNC Charlotte )
21. Acevedo J, Yan S, Michael WM*. 2016. Direct binding of RPA-coated ssDNA allows recruitment of the ATR activator TopBP1 to sites of DNA damage. Journal of Biological Chemistry. 291 (25): 13124-13131. (PMCID: PMC4933228; PMID: 27129245) (or @BioRxiv
>>> 20. Acevedo J, Yan S, Michael WM*. 2016. Direct binding of RPA-coated ssDNA allows recruitment of the ATR activator TopBP1 to sites of DNA damage. bioRxiv. (preprint online April 23, 2016) DOI: http://dx.doi.org/10.1101/050013
19. Cupello S¶, Richardson C, and Yan S*. 2016. Cell-free Xenopus egg extracts for studying DNA damage response pathways. International Journal of Developmental Biology. 60 (7-8-9): 229-236. (PMCID: PMC5071109; PMID: 27160070) DOI: http://dx.doi.org/10.1387/ijdb.160113sy
2010-2015
18. Yan S*. 2015. Teaching and learning in a Xenopus research lab. Lab Animal. 44 (8): 327. (PMCID: PMC4545573; PMID: 26200089) DOI: 10.1038/laban.817
17. DeStephanis D§,¶, McLeod M§,¶, Yan S*. 2015. REV1 is important for the ATR-Chk1 DNA damage response pathway in Xenopus egg extracts. Biochemical and Biophysical Research Communications. 460 (3): 609-615.(PMCID: PMC4426025; PMID: 25800873) DOI: http://dx.doi.org/10.1016/j.bbrc.2015.03.077
16. Richardson C*, Yan S, Vestal CG¶. 2015. Oxidative stress, bone marrow failure, and genome instability in hematopoietic stem cells. International Journal of Molecular Sciences. 16 (2): 2366-2385. (PMCID: PMC4346841; PMID: 25622253) DOI: http://dx.doi.org/10.3390/ijms16022366
15. Yan S*, Sorrell M, Berman Z¶. 2014. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cellular and Molecular Life Sciences. 71 (20): 3951-3967. (PMCID: PMC4176976; PMID: 24947324) DOI: http://dx.doi.org/10.1007/s00018-014-1666-4
14. Bai L#, Michael WM, Yan S*. 2014. Importin β-dependent nuclear import of TopBP1 in ATR-Chk1 checkpoint in Xenopus egg extracts. Cellular Signalling. 26 (5): 857-867. (PMCID: PMC3951582; PMID: 24440306) DOI: http://dx.doi.org/10.1016/j.cellsig.2014.01.006
13. Willis J§,¶, Patel Y§,‡, Lentz B ‡, Yan S*. 2013. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 110 (26): 10592-10597. (PMCID: PMC3696815; PMID: 23754435). DOI: http://dx.doi.org/10.1073/pnas.1301445110
♪ Public release by Hathaway, J. 06/14/2013. New Findings Regarding DNA Damage Checkpoint Mechanism in Oxidative Stress. (Please see the link @ UNC Charlotte and @ Eurekalert)
♪ Featured by Per, H. 2013. Biochemistry new response mechanism to oxidative stress. Biofutur. (346):17-17. (Click the link here, In French)
♪ Featured in the UNC-TV Science Week of Review: Reward, June 20, 2013 by Lane, D. (Weblink: http://science.unctv.org/content/week-review-reward)
12. Yan S*, Willis J¶. 2013. WD40-repeat protein WDR18 collaborates with TopBP1 to facilitate DNA damage checkpoint signaling. Biochemical and Biophysical Research Communications. 431 (2): 466-471. (PMCID: PMC3577970; PMID: 23333389). DOI: http://dx.doi.org/10.1016/j.bbrc.2012.12.144
♪ Highlighted on the BBRC website under Article Selections along with an introduction by Dr. Michael Lichten, BBRC Editor with expertise in DNA repair (Weblink).
11. Willis J§,¶, DeStephanis D§,¶, Patel Y‡, Gowda V‡, Yan S*.2012. Study of the DNA damage checkpoint using Xenopus egg extracts. Journal of Visualized Experiments. (69): e4449. (PMCID: PMC3514051; PMID: 23149695). DOI: http://dx.doi.org/10.3791/4449
♪ Video link:http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3514051/
10. Yan S*, Huet Y. 2011. Managing a pending IBC approval. Communicate with PI. Lab Animal. 40 (10): 295-296. (PMID: 22358201). DOI: 10.1038/laban1011-295b
♪ Featured by the Office of Laboratory Animal Welfare (OLAW) at the National Institutes of Health (NIH). (Please see the link here)
Publications before joining UNC Charlotte (total=9)
9. Van C, Yan S, Michael WM, Waga S, Cimprich KA*. 2010. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. Journal of Cell Biology. 189 (2): 233-246. (PMCID: PMC2856894; PMID: 20385778)
8. Yan S, Michael WM*. 2009. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 8 (18): 2877-2884. (PMID: 19652550)
7. Yan S, Michael WM*. 2009. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. Journal of Cell Biology. 184 (6): 793-804. (PMCID: PMC2699152; PMID: 19289795)
6. Yan S, Lindsay HD, Michael WM*. 2006. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin-bound Chk1 during checkpoint signaling. Journal of Cell Biology. 173 (2): 181-186. (PMCID: PMC2063809; PMID: 16618813)
5. Yan S*, Tso J*. 2004. Temperature may influence and regulate NF-YB expression in toad oocyte. Biochemical and Biophysical Research Communications. 313 (3): 802-811. (PMID: 14697263)
4. Yan S, Kong WH*. Gu Z, Zuo JK. 2003. Preparation and preliminary application of monoclonal antibody against cold shock domain of Y-box binding proteins. Journal of Shandong University (Natural Science Edition). 38 (4): 120-124.
3. Sun ZG, Kong WH, Yan S, Gu Z, Zuo JK*. 2003. The functions in the progesterone-induced oocyte maturation of toad ubiquitin carboxyl-terminal hydrolase (tUCH) are independent of its UCH activity. Shi Yan Sheng Wu Xue Bao (Acta Biologiae Experimentlis Sinica). 36 (2): 105-112. (PMID: 12858507)
2. Sun ZG, Kong WH, Zhang YJ, Yan S, Lu JN, Gu Z, Lin F, Tso JK*. 2002. A novel ubiquitin carboxyl terminal hydrolase is involved in toad oocyte maturation. Cell Research. 12 (3-4): 199-206. (PMID: 12296378)
1. Kong WH, Yan S, Gu Z, and Tso JK*. 2002. Developmental stage-dependence of cyclin B1 protein localization and gene expression in rabbit spermatogenesis. Sheng Li Xue Bao (Acta Physiologica Sinica). 54 (5): 400-404. (PMID: 12399820).
If you are interested in our publications, please feel free to email Dr. Yan (shan.yan@charlotte.edu).
You can also find our publications from ResearchGate: http://www.researchgate.net/profile/Shan_Yan/
Google Scholar: https://scholar.google.com/citations?hl=en&user=7i5iT5EAAAAJ
My Bibliography @NCBI: https://www.ncbi.nlm.nih.gov/myncbi/shan.yan.2/bibliography/public/
ORCID: https://orcid.org/0000-0001-8106-6295